Learning to Compare Longitudinal Images
Heejong Kim, Mert R. Sabuncu
Weill Cornell Medicine, Cornell University
Abstract
Longitudinal studies, where a series of images from the same set of individuals are acquired at different time-points, represent a popular technique for studying and characterizing temporal dynamics in biomedical applications. The classical approach for longitudinal comparison involves normalizing for nuisance variations, such as image orientation or contrast differences, via pre-processing. Statistical analysis is, in turn, conducted to detect changes of interest, either at the individual or population level. This classical approach can suffer from pre-processing issues and limitations of the statistical modeling. For example, normalizing for nuisance variation might be hard in settings where there are a lot of idiosyncratic changes.
In this paper, we present a simple machine learning-based approach that can alleviate these issues. In our approach, we train a deep learning model (called PaIRNet, for Pairwise Image Ranking Network) to compare pairs of longitudinal images, with or without supervision. In the self-supervised setup, for instance, the model is trained to temporally order the images, which requires learning to recognize time-irreversible changes. Our results from four datasets demonstrate that PaIRNet can be very effective in localizing and quantifying meaningful longitudinal changes while discounting nuisance variation.
Representative examples of datasets
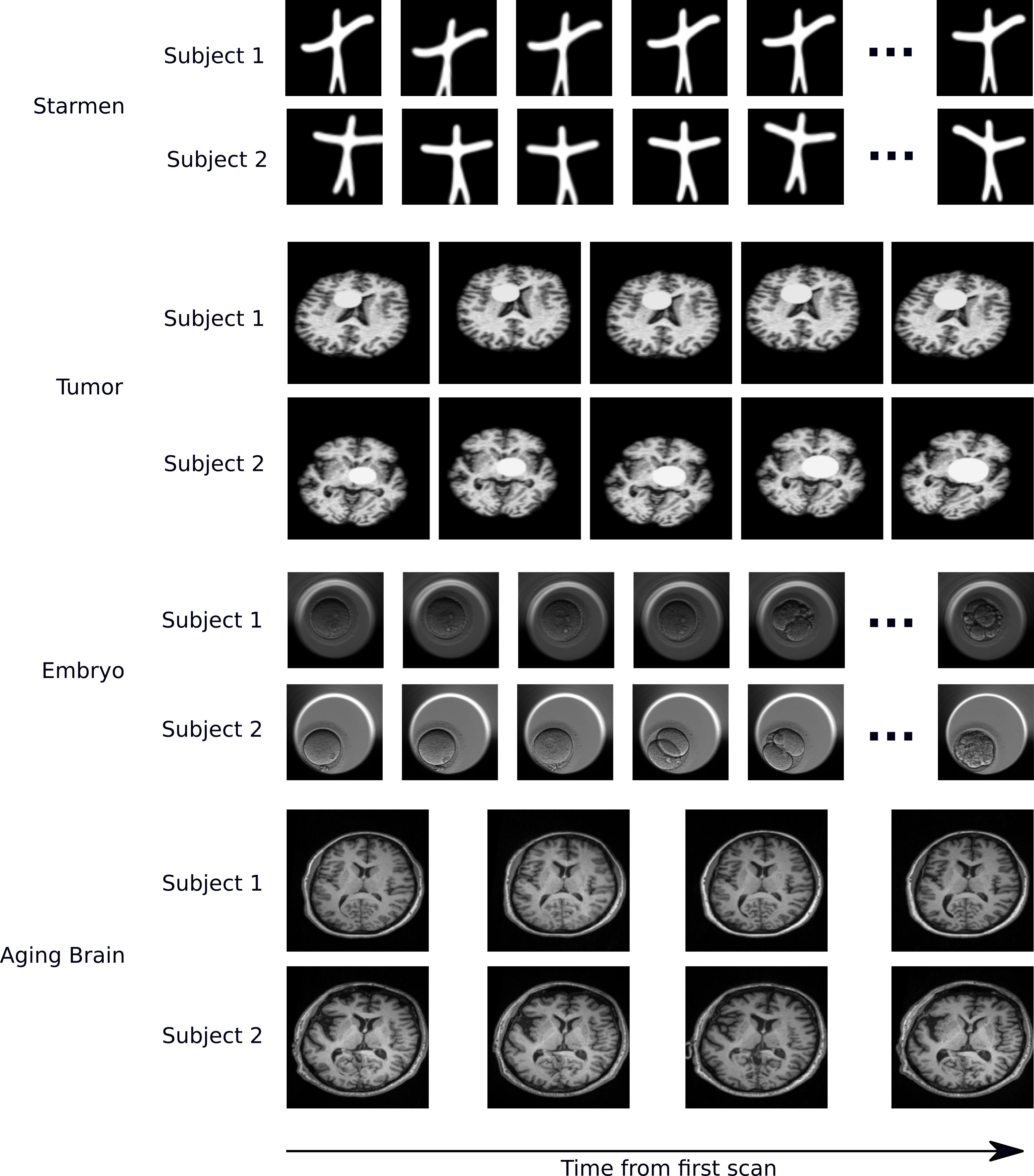
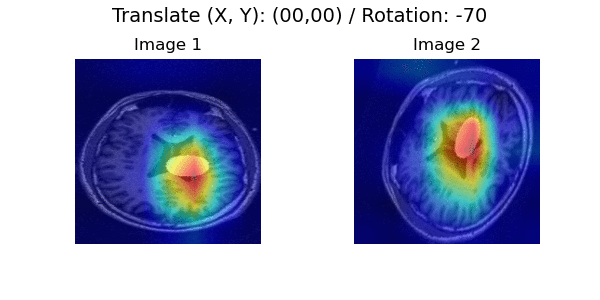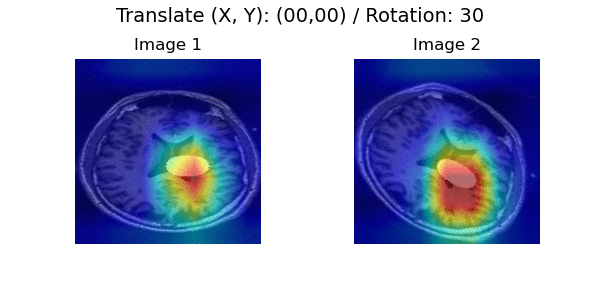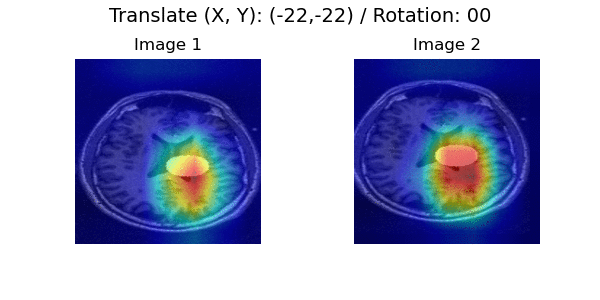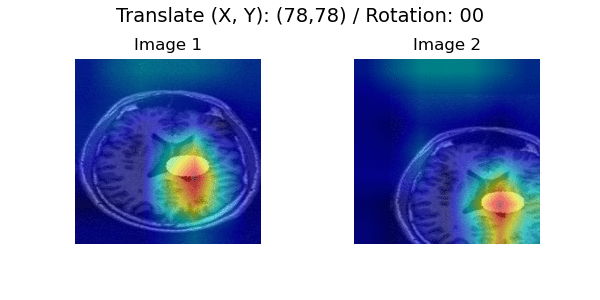
In a longitudinal imaging study, whether it is at the individual or population-level, the core challenge is disentangling nuisance variation (e.g., due to differences in imaging parameters or artifacts) from meaningful (e.g. clinically significant) temporal changes (e.g., atrophy or tumor growth).
In this paper, we present a simple learning-based approach as a novel way for analyzing longitudinal imaging data. Crucially, our approach does not require pre-processing, is model-free, and leverages population-level data to capture time-irreversible changes that are shared across individuals, while offering the ability to visualize individual-level changes.
Framework overview
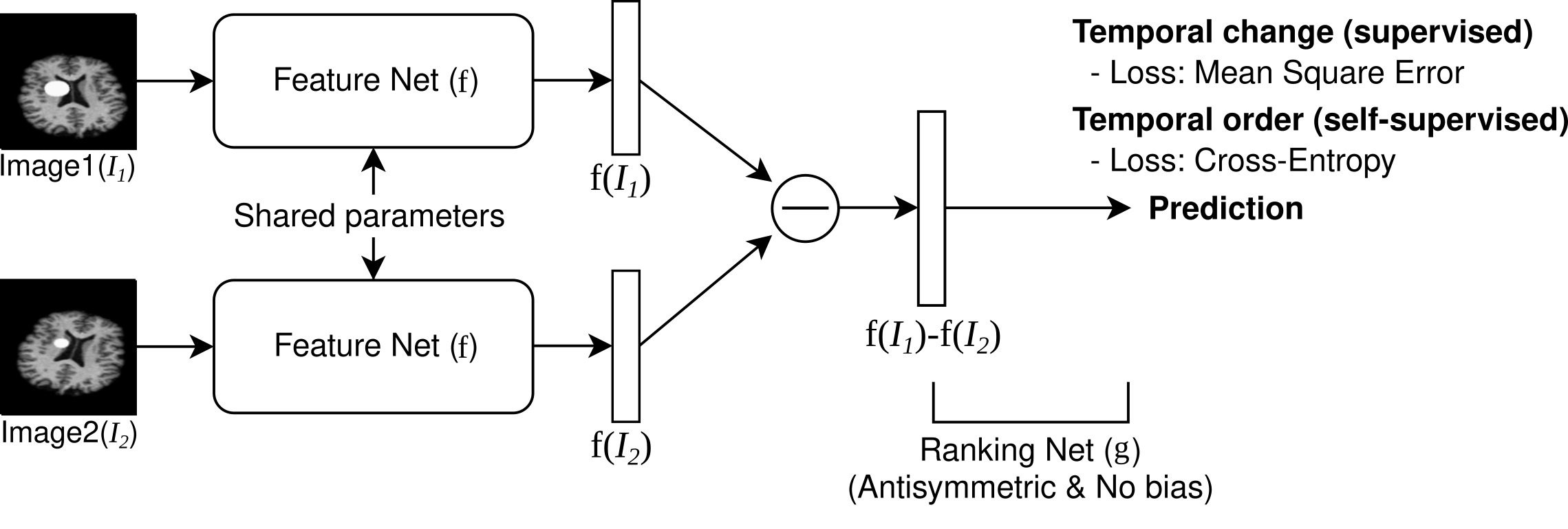
Pairwise Image Ranking Network (PaIRNet) consists of a feature extraction network, followed by a subtraction, and a ranking layer. Our core assumption is that nuisance variation, such as changes in orientation or image contrast, are independent of time, when examined across the population. We train PaIRNet to compare pairs of longitudinal images. We consider two possible tasks: 1) Supervised: PaIRNet is trained to predict a target variable that captures meaningful temporal change - such as tumor volume difference. 2) Self-supervised: the model is trained to predict the temporal ordering of the input longitudinal image pair, where the only additional information we need is the timing of the images, which is often readily available.
Result: Quantifying Longitudinal Change
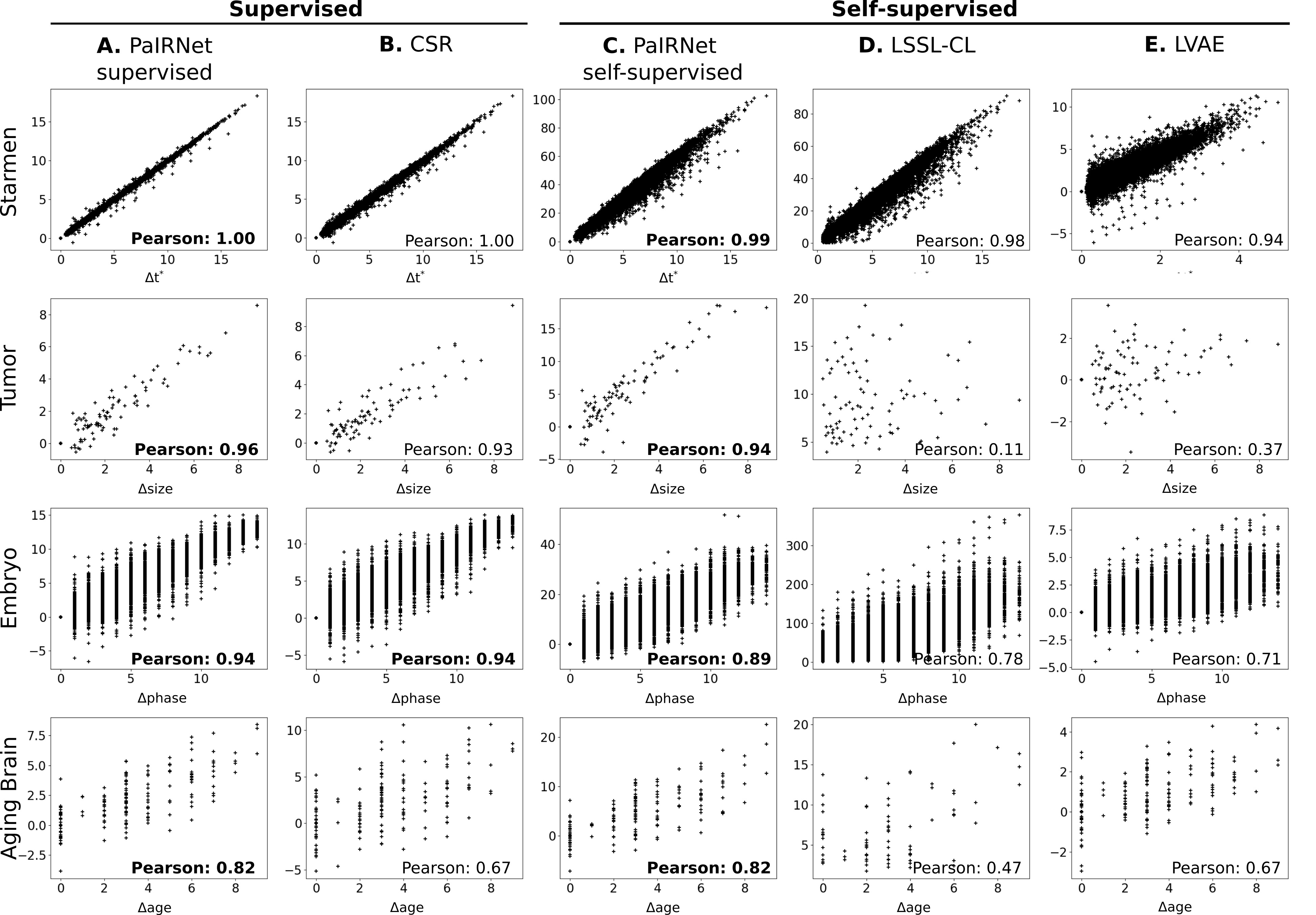
Correlation between ground-truth change in target variable and predicted change. Each row shows a different dataset and each column shows a different method. The coefficients in bold text are the best models for each task.
Overall, both supervised and self-supervised PaIRNet predictions showed strong correlation with ground-truth change. In particular, supervised PaIRNet result (A) yielded a correlation that was stronger than or equivalent to CSR (B). Self-supervised PaIRNet significantly outperformed baseline methods (LSSL-CL and LVAE).
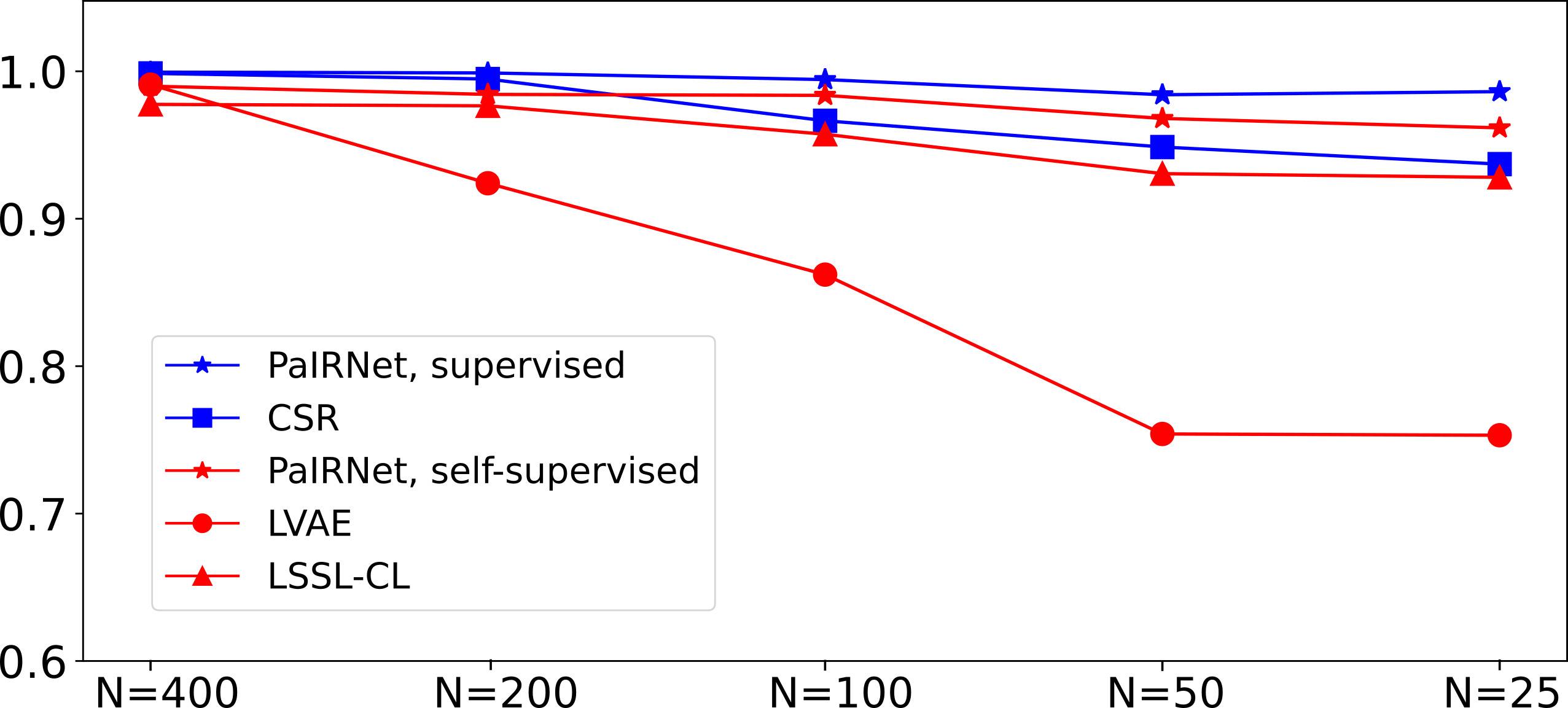
Effect of Training Data Size for Quantifying Longitudinal Change. The difference in task difficulty of pairwise comparison and CSR is highlighted in this result. We observe that supervised PaIRNet outperforms CSR with smaller training sets on the Starmen task. Correlation between ground-truth change in target variable (t*) and predicted change in models trained in the different training sizes using Starmen dataset.
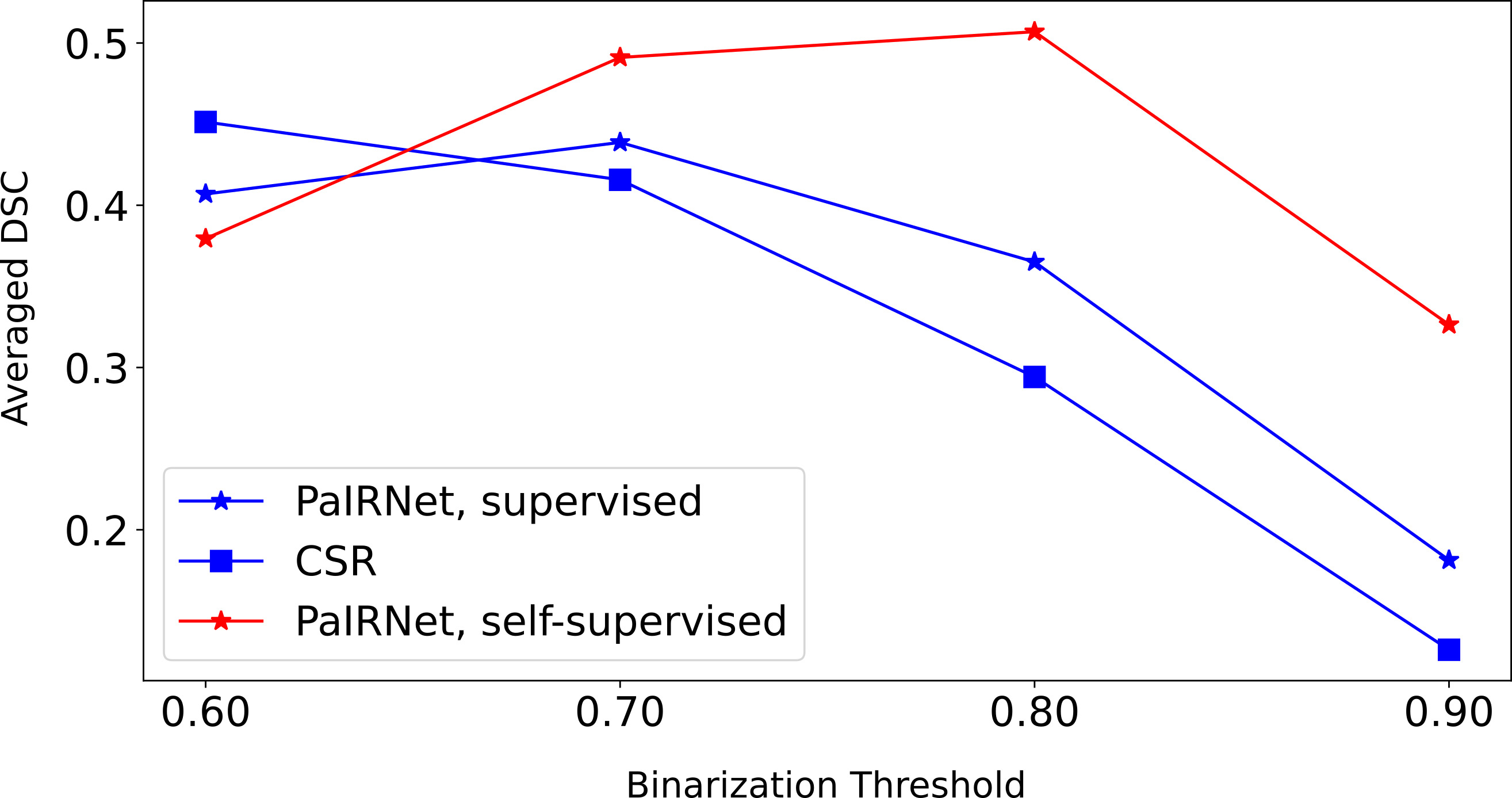
Averaged Dice Similarity Coefficient (DSC) between the target mask and the binarized activation map.
Result: Visualizing Longitudinal Change
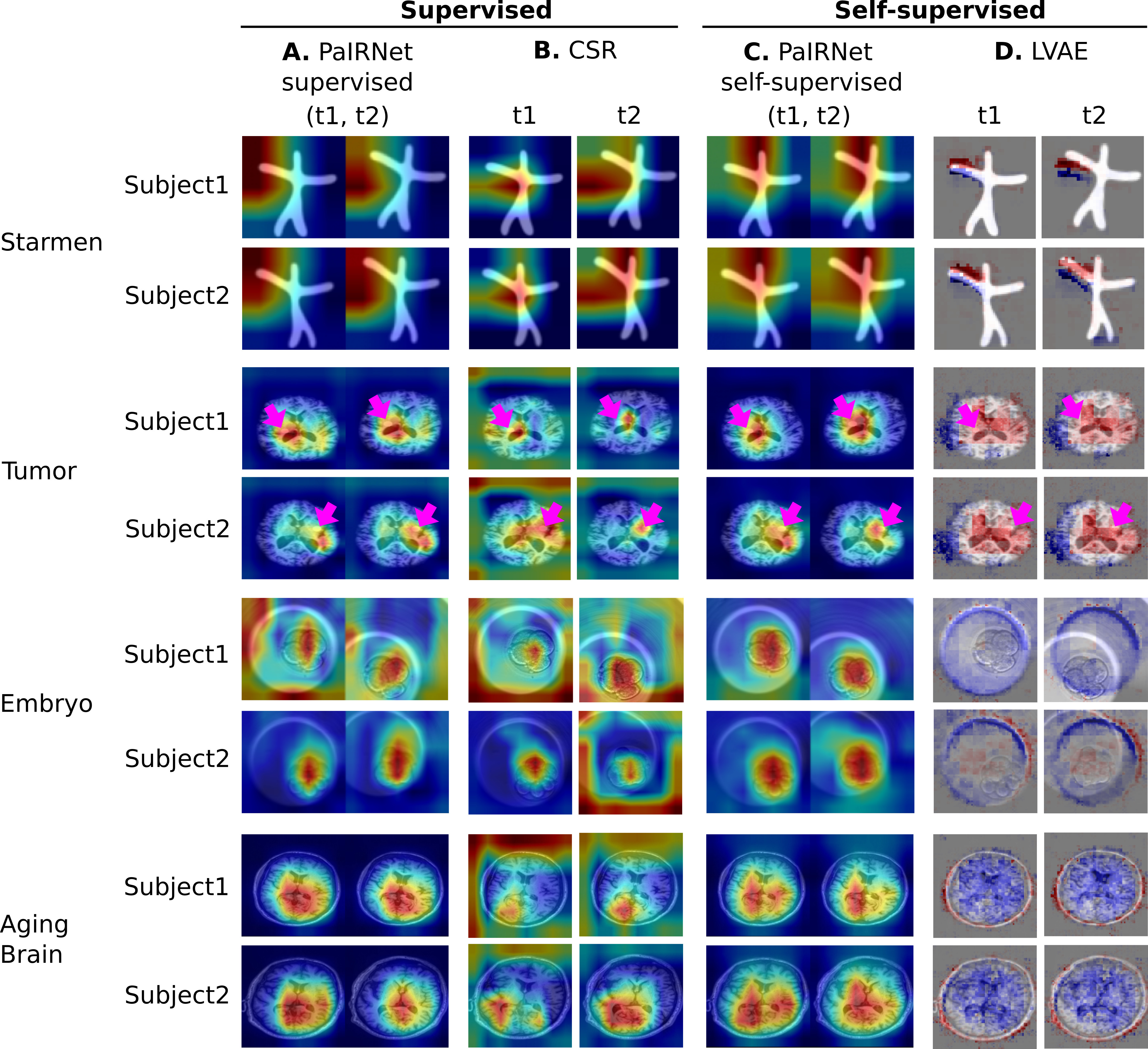
Localization results. Three time points from two subjects are visualized for each dataset. While cross-sectional supervised regression (CSR) and longitudinal variational autoencoder (LVAE) are affected by the subject-wise nuisance changes, PaIRNet discount those and locate the meaningful changes.
BibTeX
@inproceedings { kim2023learning,
title= { Learning to Compare Longitudinal Images } ,
author= { Heejong Kim and Mert R. Sabuncu } ,
booktitle= { Medical Imaging with Deep Learning } ,
year= { 2023 } ,
url= { https://openreview.net/forum?id=l17YFzXLP53 } }